Collaborative Scientific Workflow Management System (SciWorCS)
A number of Scientific Workflow Management Systems (SWfMSs) have been proposed and developed in recent years to facilitate the scientific experiments. For example some of the popular SWfMSs are: Taverna, Galaxy, Kepler, Pegasus, VisTrails, Triana, VIEW. The existing SWfMSs support only single user for a given data analysis process, however, modern scientific research projects are often collaborative in nature –involving multiple researchers of diverse domain and expertise. SciWorCS is a Collaborative Scientific Workflow management System. SciWorCS follows a plugin-based architecture for the scientific computational modules. It provides collaborative environment for scientific data analysis using efficient attribute level locking scheme. In addition to the collaborative data analysis, SciWorCS also provides real time monitoring of the computation steps.
Installation
Github link:
Please make sure you have the following environment setups:
- Python 2.7.
- Flask Mircorframework (http://flask.pocoo.org/)
- Apache CouchDB (http://couchdb.apache.org/)
Usage Instructions
On Cloning and setting up the required environment for this project, you need to follow the speps below:
1. Make sure in the project directory:
$cd SciWorCS
3. Run the main.py script as following for starting SciWorCS:
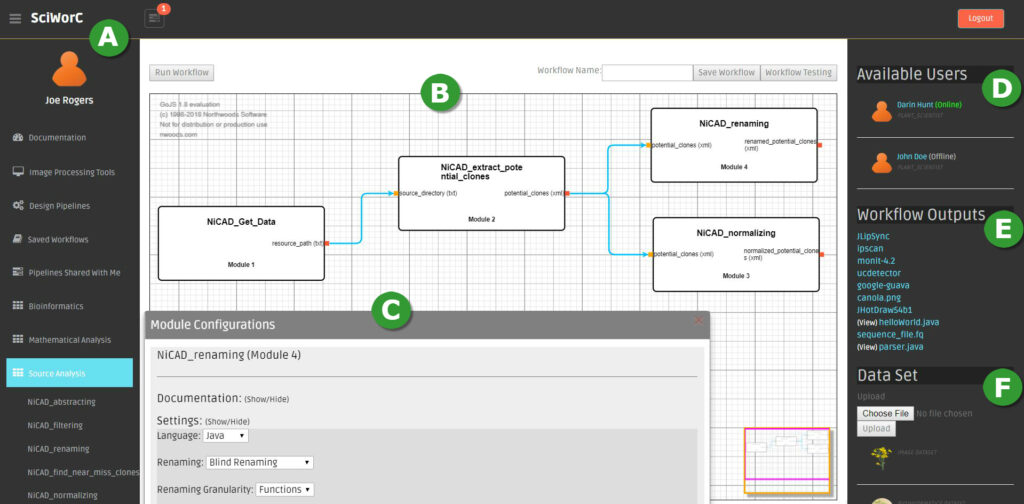
$python main.pyOn successfull installation, running the SciWorCS should display the workflow editor UI as illustrated in the following screenshot.
SciWorCS Panel A: The panel labeled as ‘A’, contains all the workflow components such as Toolbox (i.e., set of workflow modules), Saved Workflows and Shared Workflows with other users. The set of modules are classified in different Toolbox based on the general data analysis purposes of the computational modules. Some examples of such Toolboxes are Bioinformatics, Machine Learning, Source Analysis and so on as illustrated in the figure.
SciWorCS Panel B: The collaborative composition of the workflow is done on panel ‘B’. For the intended data analysis task, the required modules are selected from the corresponding Toolbox to appear in the composition panel. The selected modules are then connected together on the corresponding input/output ports for defining the datalink relation among the modules.
SciWorCS Panel C: The modules can be configured with corresponding attributes from panel ‘C’.
SciWorCS Panel D: Panel ‘D’ shows a list of collaborators and their current online/offline status.
SciWorCS Panel E: The list of the workflow outputs is shown in panel ‘E’.
SciWorCS Panel F: A new dataset can be browsed and uploaded to the central server for analysis from the panel ‘F’.
Funding Support
This work is funded by USask’s two mega CFREF (Canada First Research Excellence Fund) funded projects P2IRC and GWF. Following video demo shows how SciWorCS can be used for genotyping and phenotyping.
Papers published from this project
- Mostaeen G, Roy B, Roy C, Schneider K. (2019). Designing for Real-Time Groupware Systems to Support Complex Scientific Data Analysis. Journal Proceedings of the ACM on Human-Computer Interaction. 3(EICS): 9:1–9:28. DOI: https://doi.org/10.1145/3331151 (Journal).
- Golam M., Roy, B., Roy, C. K. and Schneider, K. A. (2018) Fine-Grained Attribute Level Locking Scheme for Collaborative Scientific Workflow Development (2018). In Proc. of IEEE Conference on Service Computing (SCC’18), pp. 273-277, doi:10.1109/SCC.2018.00047.
